The CRISPR/Cas9 (Clustered Regularly Interspaced Short Palindromic Repeats) genome-editing technology is, by itself, a significant recent addition to the biologists’ toolkit. The tool, which gives researchers the ability to direct gene edits (whether that be single point mutations, deletions of a chunk of the genome or the addition of new genetic codes) to specific sections of the genome, has seen tremendous uptake in use both in systems biology research and genetic manipulation studies.
Not only does the tool have the impact of a sledgehammer with the precision of a laser, but manipulations of the tool have resulted in a swathe of CRISPR-derived tools that either improve the tool in some specific way or alter the effect that the nuclease has on the genome being edited.
A letter published in Wiley’s Plant Biotechnology Journal in August this year described a modification of the CRIPSR tool to allow easy identification of successfully edited rice plants and simple identification of second generation plants which, while having the desired genetic modification, no longer contained the piece of transfered CRISPR/Cas9 genetic material within the genome.
The CRISPR Modification
The paper starts with the researchers noting that in the course of editing genomes in plants there are two steps that need to be successfully completed.
To edit a plant genome, the CRISPR/Cas9 genetic material needs to be transferred and inserted into the genome to be edited. This is usually performed with Agrobacterium tumefaciens which randomly inserts the CRISPR/Cas9 sequences, promoter etc into the plant genome. When the sequence is subsequently expressed, the guide RNA and the Cas9 nuclease are expressed which in turn go on to attach and edit the desired part of the genome.
For this first step to be successful, the researchers not only need to ensure that the required modification is going to be made, but must also identify the regenerated plants that have been successfully modified. Although the rate of successful editing can be quite high it is not usually 100% effective and the process of confirming a successful edit within the first generation of the modified plant can be time consuming and expensive in itself.
Once the successfully modified plants have been identified, the CRISPR/Cas9 gene insert must be removed from subsequent generations of the plant so that only the modification to the gene of interest remains. Again, this can require time consuming and expensive genetic screening processes to identify the successfully transformed, but CRISPR/Cas9 gene-free, plants.
CRISPR-S
The researchers who penned the paper theorised that it may be possible to create a gene insert that self-reported whether it was successfully introduced by causing a change in phenotype (aside from the change being introduced by the CRISPR element itself). To do this they proposed that, by introducing a gene segment into the plant that contained the CRISPR/Cas9 sequence plus a sequence that transcribed a piece of hairpin RNA complementary to some other gene of known function, the result would be the required genetic modification plus RNA interference of a marker gene that could be used for identification of successful transformation.
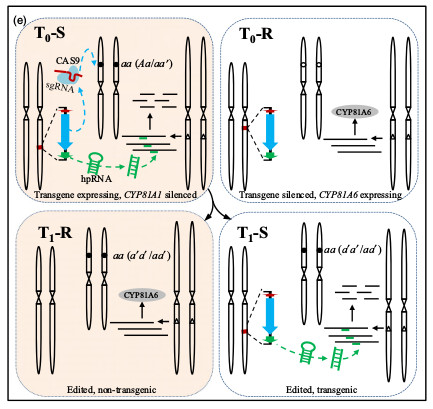
Figure 1 (e) from the article demonstrating the successful transformation of the gene in bentazon-susceptible plants (top left), the transgene-silenced and therefore resistant plants (top right), the second generation plant with the edited cadmium transport gene but lacking the CRISPR transgene, therefore regaining bentazon resistance (bottom left), and the edited second generation plant with the transgene remaining in situ (bottom right).
To demonstrate the theory they sequenced a piece of DNA to insert into rice plants. The DNA being inserted contained a CRISPR/Cas9 segment which directed an edit to a gene which encoded a cadmium transporter, plus a segment of DNA encoding a piece of hairpin RNA that, if successfully inserted into the genome of the plant, would result in the plant being susceptible to the herbicide bentazon. Bentazon susceptibility would then be the marker for successful transformation of the first generation of plants (which would assume successful editing of the gene for cadmium transportation) and also serve as a marker for those second generation plants that had lost the CRISPR gene segment given that the lack of presence of that gene would then restore the plant’s resistance to bentazon.
Testing the theory
The researchers created the sequence and inserted it into japonica rice genotype Xidao #1 via Agrobacterium tumefaciens. The tillers of the first generation of plants were divided and grown as two subplants, one of which was sprayed with bentazon, the other which wasn’t (meaning that if one subplant was susceptible to the herbicide and died, thus confirming successful transmission, the other subplant also with the successful transformation remained). Of the 96 independent transgenic events, 29 subplants were susceptible to the herbicide with 67 subplants unaffected.

Figure 1 (b) from the article demonstrating the bentazon resistant (and therefore unedited) first generation plant (left) and susceptible (and therefore edited) first generation plant (right).
The 29 subplants with the RNAi affecting bentazon resistance were then sequenced at the site of the cadmium transporter gene to test whether the gene segment had not only created the hairpin RNA but had also edited this particular gene. The researchers found that all of the plants displaying the susceptibility marker also contained the desired gene edits at the cadmium transporter gene.
Thirdly, the researchers used quantitative real-time PCR to test the abundance of the Cas9 and the bentazon susceptibility genes to check whether it was their gene construct that was causing the changes being noted. They found that in the susceptible offspring the transcripts of the Cas9 gene was significantly higher and transcripts of the gene that normally confers resistance to bentazon was significantly lower (causing the susceptibility to the herbicide) than those found in the resistant offspring. Thus, it would seem that the gene construct had successfully resulted in transcription of the Cas9 nuclease and the RNAi construct, the latter reducing the transcript abundance of the bentazon resistance gene.
To test whether their marker construct could also be used to distinguish transgene-free second generation rice plants they created 16 lines (72 seedling each) from the successfully edited first generation plants. They again sprayed the plants with bentazon with the hypothesis that those generations which had lost the transfer DNA containing the CRISPR/Cas9 and RNAi gene segment would regain their resistance to the herbicide. Of the plants that died, subsequent analysis proved that the transgene DNA remained in the genome, while the resistant plants had lost the transgene segment in the recombination of genes during the creation of the second generation.
Conclusion
It appears that these researchers have created a method, combining CRISPR/Cas9 and RNAi, to enable easy identification of successfully transformed plants and to then confirm that the transgene component has been removed in subsequent generations. This is particularly useful in those countries where current legislation does not consider plants that have been edited to remove a section of DNA (as opposed to being edited to add a piece of DNA) as “genetically modified”, therefore avoiding regulatory control. Being able to easily confirm that gene deletion has occurred and then confirm that the transgene has been removed will reduce the time and cost associated with confirming the modifications using gene sequencing.
It will be interesting to see if the process can be used in other plants. The time and expense spared could see the speed of research projects increase and open up the technology for projects that lack funding to allow for repeated primer creation and PCR-based sequencing to confirm genetic alterations.

